GUI-BioPASED: Making up an Example Task
Download a modified PDB-file used in an example task
To illustrate workflow with the program we designed a small modelling where the process of parameter specification is considered step by step. PDB:1KG5 is taken as a source protein, which is actually a K142Q mutant of E.coli MutY protein. MutY is a monofunctional DNA-N-glycosylase which excises an adenine from a heteroduplex. One of the best substrates for the MutY is a DNA containing adenine - 8-oxoguanine mispair; that accounts for appreciation of this glycosylate in the light of the research of the damage induced by an oxidative stress.
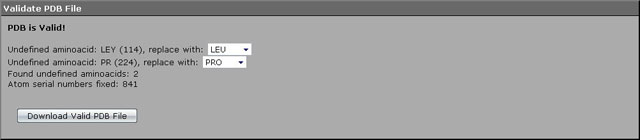
The source PDB file contains versatile information not used by the BioPASED program. In addition some errors were specially inserted to illustrate how PDB file validator works (in particular, the name of the 114th aminoacid residue - leucine - was changed to LEY; the name of the 224th aminoacid residue - proline - was changed to PR).
1. The PDB file must be first uploaded to the validation module and the valid file must be downloaded. Click the "Validate PDB File" hyperlink, then press "Browse..." and select this file in the explorer window opened (PDB file used in the task can be downloaded using the hyperlink above).
2. Then press a "Validate" button. Because of the file not containing any critical errors it passed the validation, but some unknown aminoacids were found. Select the proper aminoacid names in the drop-down combos (LEU and PRO).
3. Click the "Download Valid PDB File" button. After the download process is complete unpack the archive to some directory (e.g., "C:/ExampleModelling" in the Microsoft Windows operating system, "/home/ExampleModelling" in the Linux operating system).
After PDB file is prepared for a modelling we advance to the control files creation step.
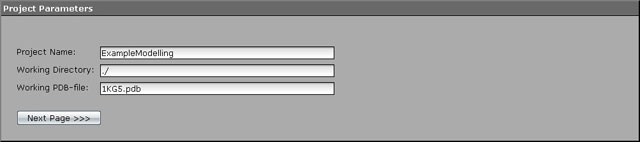
1. First general project parameters must be tweaked. Select "Project Parameters" menu item in a "Parameter groups" menu section. Enter "ExampleModelling" as the name of the project, leave a working directory intact ("./") and do not forget to specify the name of the working PDB file ("1KG5.pdb" in our task). As soon as we plan to unpack control files to the same folder as where the PDB file exist, no additional paths should be specified.
2. After filling all the fields described press the "Next Page" button to advance to the "Structure & Energy" page.
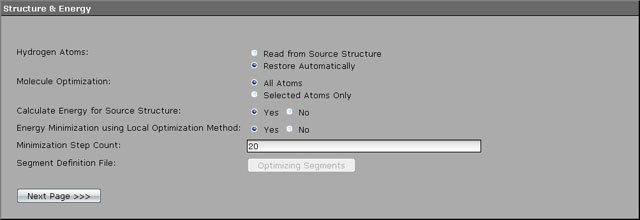
For an initial energy optimization 20 minimization steps using local descent method are performed. All we need to tweak on this page is the number of optimization steps. Enter 20 to the field named "Minimization Step Count" and advance to the next page.
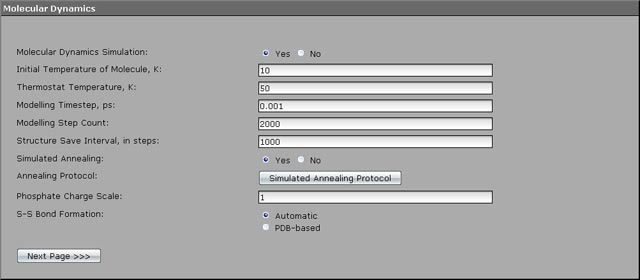
To optimize protein structure using molecular dynamics method 20000 modelling steps are performed with a timestep of 0.001 ps and structure snapshot writing every 1000 steps. The perculiarity if the modelling is a gradual warming of the molecule from 10 to 200 K using simulated annealing. We will execute warming process step by step. On the first step we will warm the molecule from 10 to 50 K for 2000 steps.
1. Enter the proper values of the parameters: 10 for the "Initial Temperature of Molecule", 50 for the "Thermostat Temperature", 2000 for the "Modelling Step Count". We leave the timestep of the default value of 0.001 ps, but the structure save interval will be provided of the value 1000. As soon as we plan to perform 20000 modelling steps (20 ps), the program will write only 20 intermediate structure snapshots.
2. The future warming we will perform using the simulated annealing method, therefore select "Yes" against the "Simulated Annealing" parameter and advance to the protocol creating by pressing the "Simulated Annealing Protocol" button.
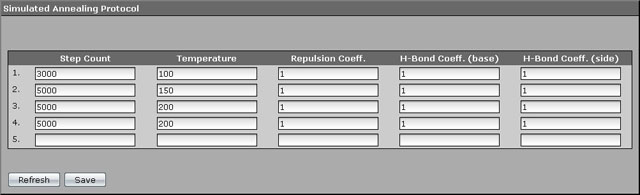
Warming of the molecule is continued gradually. We selected the following warming schema: from 50 to 100 K for 3000 steps, from 100 to 150 for 5000 steps, from 150 to 2000 for 5000 steps, and additional 5000 steps at the final value of temperature of 200 K. The filled annealing protocol will look like on the screenshot below.
3. Press "Save" and return to the "Molecular Dynamics" page.
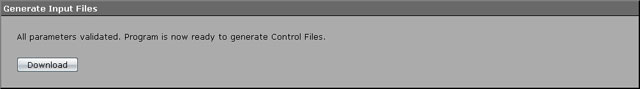
A Gaussian shell in the force field will be used as a solvation model and this is a default. Other parameters should be also left intact. Therefore you may already proceed to the creation of control files. Select "Generate Input Files" menu item. If you entered the right values of the parameters, you would be prompted to download them. Press a "Download" button, wait until download completes and unpack the archive to the directory you unpacked the valid PDB-file to.
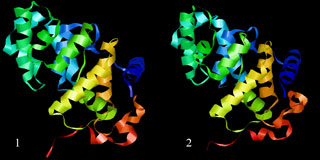
If you have installed the BioPASED program correctly, then upon launching a command file the modelling process will begin. Depending on your hardware configuration the process may take different time, at the average value of several hours. Intermediate and final structure file (ExampleModelling.mdXYZVfin.pdb) will be created in the working directory, that may be used in the necessary computations in future. The renders of the source protein molecule (1) and the modelling result (2) are shown on the image below.